Think about the number of every single microorganism you have encountered since you were born. Now add to that, all the possible microorganisms you will encounter in the future. If that sounds like a lot, you are completely correct! Now, if you remember from my previous post, I mentioned that our immune cells are able to recognize each of these microorganisms. Meaning that we have cells against influenza, cells against salmonella, cells against SARS CoV-2, etc. It seems like an impossible task to generate such a high number of diverse cells, right? I thought so too… but our body has engineered an incredible system to make this process work. Get ready to be amazed!
Receptor Recognition
As I have mentioned before, our immune system can be divided into two branches. Briefly, the first one is composed of cells that are non-specific but act very fast (known as the innate immune system) and the second of cells that are very specific but take longer to act (the adaptive immune system). Today, we will focus on the adaptive immune system.
This system is made up of two very important cell types: known as B and T cells. During an immune response, only the cells that are specific to the pathogen you are infected with should be activated. For this to occur, cells recognize a specific portion of the pathogen using a receptor. These are known as the B cell receptor (BCR) and the T cell receptor (TCR) and they are expressed on the surface of each cell.
Think about this process as a lock and a key. The interaction between the lock (the receptor on the cell) and the key (the pathogen) only occurs if they are specific to each other (you only open the lock if you have the right key). This process will give one of the signals required to activate your immune cell. Activation is essential to allow these cells to perform the functions necessary to clear the infection.
Side note, the letter O of the ImmunoThoughts logo represents a T cell (my favorite cell) and in black is the representation of a TCR. Thanks to Dr. Condotta for making this amazing logo!
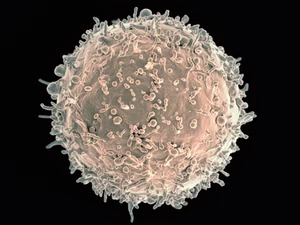
Colorized scanning electron micrograph of a B cell from a human donor. Credit: NIAID
From DNA to protein
Let’s get back to the basics for a moment. You might already know that we are made up of cells and that each cell (at least at some point during their life) has an internal manual, also known as the DNA. Think of the DNA as a recipe book. It is composed of information that allows our cells to make proteins. Proteins are in charge of doing many different tasks inside of us. They have structural, transport and storage roles. They can also participate in different chemical reactions and act as messengers sending information from one cell to another.
Therefore, to make each protein, we require a piece of information that is encoded in our DNA. For example, when a cell needs to make protein X, it will pretty much go to the “how to make protein X” part of the DNA. Now, remember about the receptors (the BRC and TCR)? They are also made up of proteins, meaning that you need a bunch of different proteins to make all the different receptor specificities (one for influenza, one for salmonella, etc).
But now think about EVERY single protein you have in your body. Lots of information required, right? Now add the huge amount of information required to make every single one of these receptors. It sounds impossible, but our body is WAY too clever! It has designed an astonishing system to “condense” all this information, and here is how…
Somatic recombination
One thing that you might not know about me is that I am bad, VERY bad, at cooking. It is not just an “it needs more salt” bad, but an “I tried… frozen pizza it is!” kinda bad. However, let’s assume that I could cook and that I woke up feeling like exploring new recipes. So, I took a blank book of 100 pages and for each page, I wrote a different ingredient. I filled the book with every single ingredient I could possibly imagine. And with that book in hand, I started my cooking adventure as following.
For my first recipe, I added the ingredients from page 1 to 4, I removed pages 5 to 22 and added the ingredients from pages 23 to 100. After I was done, I tasted my new cooking invention and asked myself “does it work?” (does it taste good?). If it didn’t work, I tried again. This time I added the ingredients from pages 1 to 4, removed extra pages (pages 23 to 35, let’s say), and added those from pages 36 to 100. Once completed, I asked myself the same question: “does it work?”. If it did, then great. But if it didn’t, I continued removing pages until I find something that worked for my taste, or until the book was over (in which case a new book would be required to repeat this process).
As insane as this idea sound, our body has evolved to utilize a similar process. A mechanism that leads to high receptor diversity while maintaining a low storage space. During the generation of B and T cells, developing cells will access the “how to make a receptor” area of the DNA and perform what is known as somatic recombination. Cells will try to generate a protein using different portions of the DNA present in this location. If the recombination works (leading a viable receptor) the cell is good to go. But if it doesn’t work, a piece of DNA will be cut (or the pages of the book will be removed) and a new protein will be generated and expressed on the surface of the cell for testing. This process will occur many times until a viable receptor is generated or until no more recombinations are possible.
Keep in mind that I am presenting this process in a very simplistic way. But, let me know if you’d like me to cover this mechanism in greater detail! The goal of this post is to explain how is possible for us to have so many different receptors using only a small area in the DNA.
This occurs because each cell will cut the DNA a bit differently. Believe it or not, this process ensures that you can generate cells that are specific to each pathogen. For example, you already have cells that are specific to a portion of the SARS CoV-2, even though you might have never been infected with it. And this is only possible due to the ability of our adaptive immune cells to undergo somatic recombination!
Self vs non-self
But wait a moment… I’m telling you that, this pretty much random process leads to cells that are able to recognize a portion of a pathogen, but how does it know what is a pathogen? How can we rule out that we are not making receptors that are specific to heart cells (for example)? GREAT thinking and the answer is… we don’t.
Somatic recombination does not rule out the possibility of generating cells that are specific to our own cells. However, the development of our adaptive immune cells is a very regulated process. There are different mechanisms that together prevent the survival of cells that could attack our own body. Unfortunately, when these “deletion” mechanisms don’t work we can develop autoimmune diseases… but I will keep this subject for my next posts!
From your immunologist – in training,
Stefanie Valbon
PS: If after reading this post you start to have a few ideas on why my cooking abilities are not very great… you might be correct… but, all for science, right!?